Dr. Izumiya Research Lab-
Yoshi Izumiya, D.V.M., Ph.D.
Professor

Department of Dermatology
UC Davis School of Medicine
Research III
4645 Second Avenue
Sacramento, CA 95817
Phone: 916-734-7842
Biography
Dr. Izumiya received his undergraduate degree in Veterinary Medicine at Kitasato University, Aomori, Japan in 1997, followed by a Ph.D. degree in Veterinary Microbiology in 2001 at the University of Tokyo, Tokyo, Japan. Between 1999 and 2001, Dr. Izumiya was a fellow of the Japanese Society for the Promotion of Science. After Dr. Izumiya received his Ph.D. degree, he moved to the U.S. and expanded his research from animals to human as a postdoctoral fellow in School of Medicine at UC Davis. As a postdoctoral fellow, he received an award for Excellence in Postdoctoral Research from UC Davis. In 2005, Dr. Izumiya was promoted to the title of Assistant Research Biochemist in the Department of Biochemistry and Molecular Medicine. In 2009, Dr. Izumiya joined the UC Davis Department of Dermatology and was promoted to Assistant Professor.
In his time at UC Davis, Dr. Izumiya was awarded multiple research funds including a postdoctoral fellowship and IDEA award from California HIV/AIDS Research Program, Health Science System Award as well as Translational Research Grant from the CTSC. Other research supports includes grants from USDA and NIH. He also serves as a reviewer for virology related journals such as the Journal of Virology, Virology and NIH Viral B study section.

 |
Yoshihiro Izumiya, D.V.M., Ph.D. Associate Professor Research interests: Gene regulation of viruses such as Kaposi’s Sarcoma-Associated Herpesvirus |
 |
Mel Cambell, Ph.D. Assistant Project Scientist |
 |
Chie Izumiya, D.V.M. Assistant Project Scientist |
|
Kazushi Nakano, Ph.D. |
|
 |
Kevin Kim |
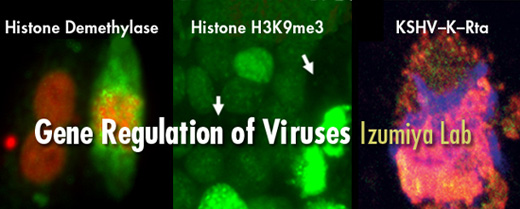
Yoshihiro Izumiya D.V.M. Ph.D. and his laboratory study epigenetic gene regulation by using herpesvirus reactivation as a model system. Many viruses produce an acute infection in the human host and are then cleared by the immune system. However, other viruses, especially herpesvirus, remain in the infected person till death by producing a chronic latent infection. In latent infection, the herpesvirus hides from the immune system by silencing their own gene expression and no infectious virus particles can be detected during latency. The virus can be reactivated with different external stimuli and will then activate hundreds of gene expression; this provides us a compelling model to study movement and regulation from “silenced chromatin” to “active chromatin” of host genome. In addition, very long history of co-evolution with host also makes viral protein as an attractive starting material for drug development to regulate host protein function, because viral replication absolutely relies on host cell machineries. Viruses are very smart to take over the host cell protein functions and we are hoping to reverse-hijack by studying viral-host protein interactions.
Dr. Izumiya’s lab is particularly interested in the epigenetic components of chromatin, including host and viral factors that interact to facilitate both the establishment and maintenance of the chromatin state as well as the molecular transitions that take place during reactivation (“chromatin remodeling”). In addition, developing anti-cancer drugs in a form of antibody-conjugate to deliver small peptide identified from virology is also an area of research in his lab. This is done by a collaboration with a start-up company, VGN. Bio, Inc. Moreover, their research may shed new information applicable to other areas of biology, such as cancer, development and aging, where epigenetic regulation has been implicated.
Progress Reports (2018-2022)
- Kumar A, Lyu Y, Yanagihashi Y, Chantarasrivong C, Majerciak V, Salemi M, Wang KH, Chuang F, Davis RR, Tepper CG, Nakano K, Izumiya C, Shimoda M, Nakajima KI, Merleev A, Zheng ZM, Campbell M, & Izumiya Y* (2022) KSHV episome tethering sites on host chromosomes and regulation of latency-lytic switch by CHD4. Cell Reports 39(6):110788. PMID: 35545047
- Merleev A, Le ST, Alexanian C, Toussi A, Xie Y, Marusina AI, Watkins SM, Patel F, Billi AC, Wiedemann J, Izumiya Y, Kumar A, Uppala R, Kahlenberg JM, Liu FT, Adamopoulos IE, Wang EA, Ma C, Cheng MY, Xiong H, Kirane A, Luxardi G, Andersen B, Tsoi LC, Lebrilla CB, Gudjonsson JE, Maverakis E (2022) Biogeographic and disease-specific alterations in epidermal lipid composition and single cell analysis of acral keratinocytes. JCI Insight PMID: 35900871
- Campbell M, Chantarasrivong C, Yanagihashi Y, Inagaki T, Davis RR, Nakano K, Kumar A, Tepper CG, & Izumiya Y* (2022) KSHV Topologically Associating Domains in Latent and Reactivated Viral Chromatin. Journal of Virology 96(14): e0056522. PMID: 35867573
- Merleev A, Ji-Xu A, Toussi A, Tsoi LC, Le ST, Luxardi G, Xing X, Wasikowski R, Liakos W, Brüggen MC, Elder JT, Adamopoulos IE, Izumiya Y, Riera-Leal A, Li Q, Kuzminykh NY, Kirane A, Marusina AI, Gudjonsson JE, Maverakis E (2022) Proprotein convertase subtilisin/kexin type 9 (PCSK9) is a psoriasis susceptibility locus that is negatively related to IL36G. JCI Insight PMID: 35862195
- Shimoda M, Lyu Y, Wang KH, Kumar A, Miura H, Meckler JF, Davis RR, Chantarasrivong C, Izumiya C, Tepper CG, Nakajima KI, Tuscano J, Barisone G, & Izumiya Y* (2021) KSHV transactivator-derived small peptide traps coactivators to attenuate MYC and inhibits leukemia and lymphoma cell growth. Communications Biology 4(1): 1330 PMID: 34857874
- Kumar A, Salemi M, Bhullar R, Guevara-Plunkett S, Lyu Y, Wang KH, Izumiya C, Campbell M, Nakajima KI, & Izumiya Y* (2021) Proximity Biotin Labeling Reveals Kaposi’s Sarcoma-Associated Herpesvirus Interferon Regulatory Factor Networks. Journal of Virology 95 (9): e02049-20 PMID: 33597212
- Liao Y, Lupiani B, Izumiya Y, & Reddy SM (2021) Marek’s disease virus Meq oncoprotein interacts with chicken HDAC 1 and 2 and mediates their degradation via proteasome dependent pathway. Scientific Reports 1; 637 PMID: 33437016
- Nakajima KI, Guevara-Plunkett S, Chuang F, Wang KH, Lyu Y, Kumar A, Luxardi G, Izumiya C, Soulika A, Campbell M, & Izumiya Y*. (2020) Rainbow-KSHV Revealed Heterogenic Replication with Dynamic Gene Expression. Journal of Virology 94 (8). e01565-19 PMID: 31969436
- Campbell M, & Izumiya Y*. (2020) PAN RNA: Transcription exhaust from a viral engine. Journal of Biomedical Science 27 (1): 41 PMID: 32143650
- Liao Y, Lupiani B, Bajwa K, Khan OA, Izumiya Y, & Reddy SM (2020) Role of Marek’s Disease Virus (MDV)-Encoded US3 Serine/Threonine Protein Kinase in Regulating MDV Meq and Cellular CREB Phosphorylation. Journal of Virology 94 (17): e00892-20 PMID: 32581093
- Zhou F, Liu X, Gao L, Zhou X, Cao Q, Niu L, Wang J, Zuo D, Li X, Yang Y, Hu M, Yu Y, Tang R, Lee BH, Choi BW, Wang Y, Izumiya Y, Xue M, Zheng K, & Gao D. (2019) HIV-1 Tat enhances purinergic P2Y4 receptor signaling to mediate inflammatory cytokine production and neuronal damage via PI3K/Akt and ERK MAPK pathways. Journal of Neuroinflammation 16:71 PMID: 30947729
- Chen CP, Chuang F, Izumiya Y*. Functional Imaging of Viral Transcription Factories Using 3D Fluorescence Microscopy. J Vis Exp. 2018 Jan 18;(131 PMID: 29443057
- Campbell M, Watanabe T, Nakano K, Davis RR, Lyu Y, Tepper CG, Durbin-Johnson B, Fujimuro M, Izumiya Y*. KSHV episomes reveal dynamic chromatin loop formation with domain-specific gene regulation. Nat Communs. 2018 Jan 4;9(1):49. PMID: 29302027
Book Chapter:
Davis RR, Campbell M, Izumiya Y, & Tepper CG (2020) Capture Hi-C: Characterization of Chromatin Contacts. Epigenetic Methods, Chapter 21
